System Biosciences
EF1a-hsaCas9-U6-gRNA(SA) linearized all-in-one SmartNuclease AAV Plasmid
- SKU:
- CASAAV100PA
- Availability:
- Usually Shipped in 5 Working Days
- Size:
- 10 rxn
- Shipping Temperature:
- Blue Ice
Description
EF1a-hsaCas9-U6-gRNA(SA) linearized all-in-one SmartNuclease AAV Plasmid. Cat# CASAAV100PA. Supplier: SBI System Biosciences

Overview

- Deliver Cas9 in vivo
- Edit genomes in post-natal animals
- Develop gene therapies in small animal models
- Generate novel disease models
- Choose from All-in-one or Two Vector AAV-Cas9 systems


How It Works
Using saCas9
While saCas9 is just as efficient as spCas9, a few differences between the two systems will affect gRNA design1.
- saCas9 PAM differs from spCas9 PAM
- saCas9 works most efficiently with gRNAs of 21 nt – 23 nt
Contact us with any questions on gRNA design for saCas9 or about using our AAV-Cas9 vectors by emailing tech@systembio.com.
saCas9 PAM sequences:
NNGGGT
NNGAAT
NNGAGT
To create gRNAs for AAV Cas9 SmartNuclease Plasmids, use the following:
Fwd-5.1: ACCGNNNNNNNNNNNNNNNNNNNNN (N target sequence or gRNA sequence, 21 nts)
Rev-3.1: aaacXXXXXXXXXXXXXXXXXXXXX (X reverse complementary of N, 21 nts)
Example:
aavs1 gRNA for saCas9: CTGTCCCTAGTGGCCCCACTG
sa-AAVS1gRNA-5.1 ACCGCTGTCCCTAGTGGCCCCACTG
sa-AAVS1gRNA-3.1 aaacCAGTGGGGCCACTAGGGACAG
The workflow at-a-glance
- Design two DNA oligonucleotides that are sense and antisense sequences of the target DNA and are immediately upstream of a PAM sequence (see above section on creating gRNAs for AAV Cas9 SmartNuclease Plasmids)
- Anneal the two oligonucleotides to generate a duplex
- Ligate the duplex into the pre-linearized All-in-one AAV Cas9 SmartNuclease Plasmid
- Transform into competent cells and grow in LB/Kanamycin plate (50 µg/ml)
- Confirm positive clones by direct sequencing
- Transfect sequence-verified All-in-one construct into AAV packaging cells
- Isolate packaged All-in-one AAV Cas9 SmartNuclease Plasmid using AAVanced AAV Concentration Reagent for easy, high-titer preparations
Your AAV Cas9/gRNA virus is ready for your genome editing project.
Genome engineering with CRISPR/Cas9
For general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous
recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system
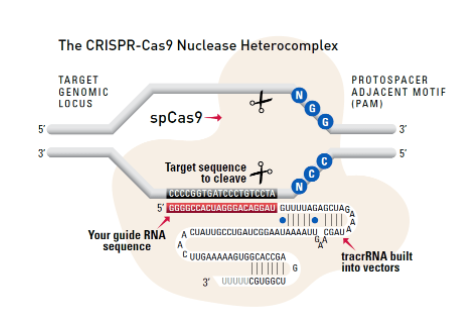
Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor
plasmid determines whether the homologous recombination event results in a knock-out,
knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR
donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells
using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.
Supporting Data
Genome engineering with SBI’s All-in-one AAV Cas9 SmartNuclease Plasmids
Both SBI’s all-in-one AAV-Cas9 and two-vector AAV-Cas9 systems efficiently correct a dysfunctional genomic copy of EGFP.
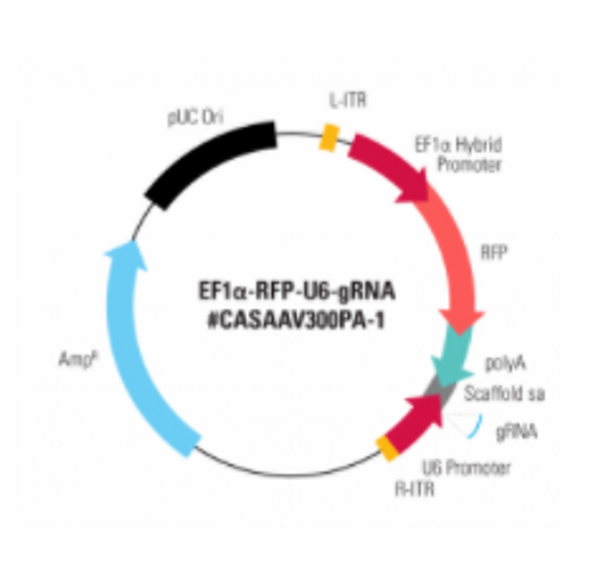
Starting with an Enhanced Green Fluorescent Inhibited Protein (EGIP) reporter cell line, which has an inactivated EGFP due to a premature stop codon, we designed a gRNA sequence and HR rescue donor to remove the stop codon and enable GFP expression. The gRNA sequence was inserted into either Cat # CASAAV300PA-1, EF1-RFP-U6-gRNA(SA)—the gRNA-expressing plasmid in the Two Vector AAV Cas9 SmartNuclease System (Figure 1, top panels)—or Cat # CASAAV100PA-1, the EF1α-hsaCas9-U6-gRNA(SA) All-in-one AAV Cas9 SmartNuclease Plasmid (Figure 1, bottom panels).
AAV virus particles isolated from both cell lysates (Figure 1, left panels) and packaging supernatants (Figure 1, right panels) were delivered to cells with the rescue HR donor and restored EGFP expression, indicating successful genome editing.
Delivery of AAV plasmid(s) alone (Figure 1, middle panels) with HR donor also restored EGFP expression, again indicating successful genome editing.
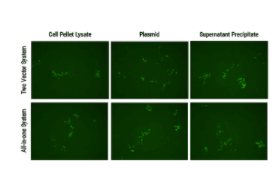
Figure 1. Both SBI’s all-in-one AAV-Cas9 and two-vector AAV-Cas9 systems efficiently correct a dysfunctional genomic copy of EGFP. METHODS: Approximately 1 – 1.5 x 106 EGIP reporter cells were seeded into a 12-well plate for infection with isolated AAV particles (left and right panels) or transfection with plasmid (middle panels), along with the HR rescue donor. The top panels were treated with the Two Vector AAV Cas9 SmartNuclease System plus HR rescue donor designed to correct the defect in EGIP, restoring EGFP expression. The bottom panels were treated with the All-in-one AAV Cas9 SmartNuclease Plasmid plus the same HR rescue donor. Three days post-infection or transfection, EGFP-positive cells can clearly be seen, indicating successful genome editing.