System Biosciences
CLOuD9 Inducer Reagent (0.5M) - 400 ul
- SKU:
- CASCL9-200A
- Availability:
- Usually Shipped in 5 Working Days
- Size:
- 200 ul
- Shipping Temperature:
- Blue Ice
Description
CLOuD9 Inducer Reagent (0.5M) - 400 ul. Cat# CASCL9-200A. Supplier: SBI System Biosciences

Overview
Already included in SBI’s exclusive CLOuD9 Gene Expression Regulation Kit, CLOuD9 Inducer Reagent is also available separately from the kit. Learn more about the CLOuD9 Gene Expression Regulation Kit—introduced in a recent Nature Communications paper1—by visiting the product page.
CLOuD9 for Chromatin Loop Reorganization using CRISPR-dCas9:
- Innovative—leverage the simplicity of CRISPR/Cas9 targeting—free of DNA cleavage activity—for inducible and reversible chromatin loop formation anywhere in the genome
- Well-validated—chromatin loop formation and the resulting changes in gene expression across multiple loci and cell lines is amply demonstrated in Morgan, et al,1
- Broadly applicable—great for studying the role of chromatin in the control of gene expression in a range of areas, including:
- Developmental biology
- Cancer biology
- Chromatin structural biology
- Chromatin-based therapeutic strategies
- Unique—the innovative CLOuD9 System is only sold by SBI
- Morgan, SL, et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun. 2017. Jul 13; 8:15993. PMCID: PMC5511349.
How It Works
Form chromatin loops with CLOuD9
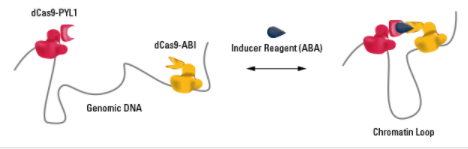
Figure 1. The CLOuD9 System enables formation of inducible and reversible chromatin loops anywhere in the genome.
The CLOuD9 System consists of three components (Figure 1):
- A null mutant of the Cas9 protein from aureus fused to the plant ABI protein
- A null mutant of the Cas9 protein from pylogenes fused to the plant PYL1 protein
- Inducer reagent, the plant phytohormone S-(+)-abscisic acid (ABA)
The use of the Cas9 null mutants enables accurate binding to a specific region of the genome via the user-designed guide RNA (gRNA) without generating any breaks in the DNA. Because the gRNA scaffolds from the two species bind more efficiently to their same-species Cas9 partner, we can direct the saCas9-ABI fusion localizes to one region of the DNA and the spCas9-PYL1 fusion to a second region of DNA.
The dimerization partners—ABI and PYL1—are part of the plant ABA signaling pathway, and form heterodimers in the presence of ABA. Dimerization of ABI and PYL1 is reversible, enabling formation of chromatin loops in the presence of ABA, and disassembly of the chromatin loops upon ABA washout.
Supporting Data
Study the role of chromatin structure in gene expression
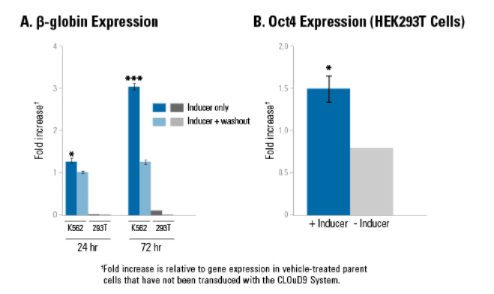
Figure 2. CLOuD9-induced chromatin loops increase gene expression in a context-dependent manner.
1. A CLOuD9-induced chromatin loop at the β-globin locus leads to activation of gene expression in K562 cells but not in HEK293T cells, highlighting the importance of cellular context and the need for additional factors in gene expression. Inducer reagent (in DMSO) was added for either 24-hours or 72-hours. At the indicated time, cells in the “inducer only” treatment group were harvested and analyzed for gene expression, whereas cells in the “inducer + washout” treatment group were washed and then allowed to grow another 24- or 72-hours as indicated in the absence of inducer. Significance assessed relative to DMSO-treated control cells which were not transduced with the CLOuD9 System. * indicates P < 0.05, *** indicates P < 0.0001.
2. Unlike the β-globin locus, a CLOuD9-induced chromatin loop (treated with Inducer Reagent in DMSO) results in robust expression of the Oct4 gene in CLOud9/HEK293T cells compared to CLOuD9/HEK293T cells treated with DMSO only. The CLOuD9 System fusion proteins were targeted to the Oct4 promoter and the 5’ distal enhancer region upstream of the Oct4 promoter. *indicates P < 0.05.