System Biosciences
Extra T7 gRNA PCR primer mix (5 uM)
- SKU:
- CAS510A-PR
- Availability:
- Usually Shipped in 5 Working Days
- Size:
- 50 assays
- Shipping Temperature:
- Blue Ice
Description
Extra T7 gRNA PCR primer mix (5 uM). Cat# CAS510A-PR. Supplier: SBI System Biosciences
Overview

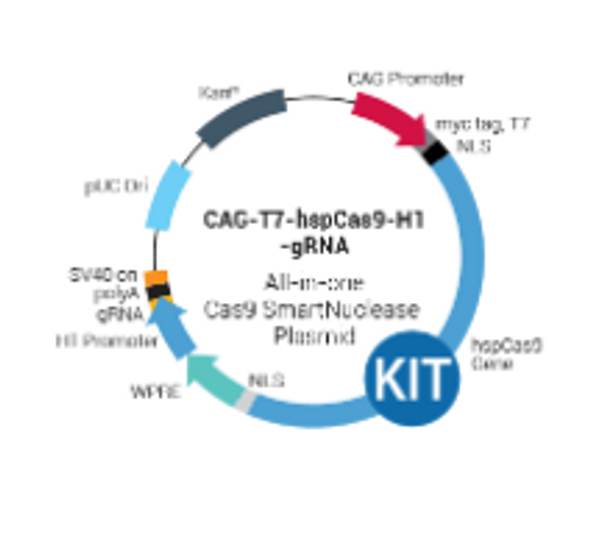
How It Works
Synthesizing gRNA
The workflow at-a-glance
- Design two DNA oligonucleotides that are sense and antisense sequences of the target DNA and are immediately upstream of a PAM sequence (5’ – NGG – 3’)
- Anneal the two oligonucleotides to generate a duplex
- Ligate the duplex into the pre-linearized T7 gRNA SmartNuclease Cloning and Production Vector
- Transform into competent cells and grow in LB/Kanamycin plate (50 µg/ml)
- Confirm positive clones by direct sequencing
- Perform in vitro transcription (IVT) with the T7 SmartNuclease™ gRNA Synthesis Kit (Cat.# CAS510A-KIT)
Selecting Target DNA Sequences
The selection of the target DNA sequence is not limited by any constraints, with exception of the requirement of a PAM sequence in the form of 5’ – NGG – 3’ (where N = any base) immediately following the target sequence. The typical length of the target sequence is 20 bp.
Genome Engineering
For more general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous
recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system

Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor
plasmid determines whether the homologous recombination event results in a knock-out,
knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR
donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells
using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.
Supporting Data
The T7 gRNA SmartNuclease Cloning and Production Vector in action

Figure 1. gRNA cloned-into and synthesized using the T7 gRNA SmartNuclease Cloning and Production Vector directs rescue of a non-fluorescent eGFP mutant using Purified Cas9 Protein. Rescue of non-fluorescent eGFP mutant (EGIP) via homology-directed repair using either a Cas9 All-in-one plasmid system (left panel) or a Cas9 protein-gRNA system (right panel), three days post-transfection. Direct transfection of Cas9 protein-gRNA results in a higher rescue efficiency.