System Biosciences
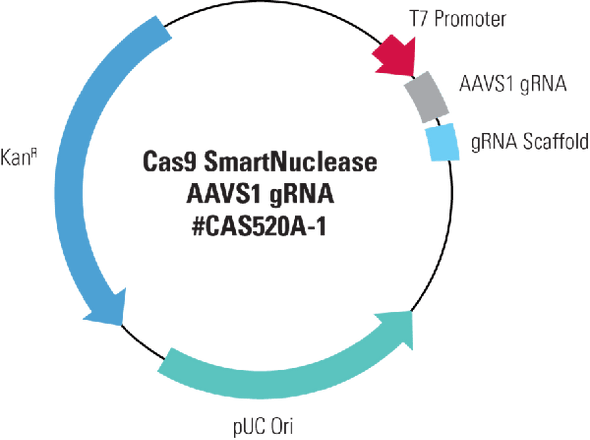
Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter]
- SKU:
- CAS601A
- Availability:
- Usually Shipped in 5 Working Days
- Size:
- 10 ug
- Shipping Temperature:
- Blue Ice
Description
Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter]. Cat# CAS601A. Supplier: SBI System Biosciences

Overview

- Easy, precise knock-in of any gene
- Consistent, robust transgene expression from the AAVS1 Safe Harbor Site
- Simplified construction of isogenic cell lines
- Minimal off-target integration
- Streamlined CRISPR/Cas9 library screening
How It Works
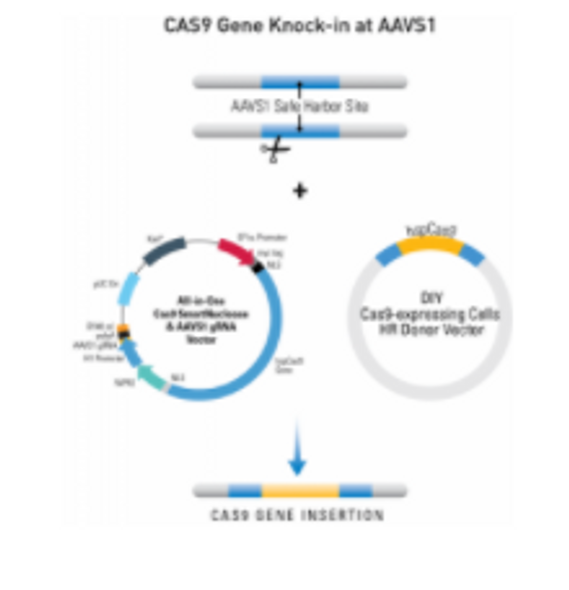
Gene knock-in at AAVS1

Figure 1. Knocking-in a gene at the AAVS1 site using an HR Targeting Vector. Step 1: Cas9 creates a double-stranded break(DSB) at the AAVS1 site. Cas9 activity is directed to the AAVS1 site by an AAVS1-specific gRNA. Step 2: The DNA repair machinery is recruited to the DSB. In the presence of an HR Donor with homology to the region adjacent to the DSB (blue areas of the genomic and plasmid DNA) homologous recombination (HR) is favored over non-homologous end joining (NHEJ). Result: The HR event leads to insertion of the region of the HR Donor Vector between the two homology arms—your gene-of-interest is integrated into the AAVS1 site.
Genome engineering with CRISPR/Cas9
For general guidance on using CRISPR/Cas9 technology for genome engineering, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »
CRISPR/Cas9 Basics
Through careful selection of the target sequence and design of a donor plasmid for homologous
recombination, you can achieve efficient and highly targeted genomic modification with CRISPR/Cas9.
The system

Cas9 protein—uses guide RNA (gRNA) to direct site-specific, double-strand DNA cleavage adjacent to a protospacer adapter motif (PAM) in the target DNA.
gRNA—RNA sequence that guides Cas9 to cleave a homologous region in the target genome. Efficient cleavage only where the gRNA homology is adjacent to a PAM.
PAM—protospacer adapter motif, NGG, is a target DNA sequence that spCas9 will cut upstream from if directed to by the gRNA.
The workflow at-a-glance
DESIGN: Select gRNA and HR donor plasmids. Choice of gRNA site and design of donor
plasmid determines whether the homologous recombination event results in a knock-out,
knock-in, edit, or tagging.
CONSTRUCT: Clone gRNA into all-in-one Cas9 vector. Clone 5’ and 3’ homology arms into HR
donor plasmid. If creating a knock-in, clone desired gene into HR donor.
CO-TRANSFECT or CO-INJECT: Introduce Cas9, gRNA, and HR Donors into the target cells
using co-transfection for plasmids, co-transduction for lentivirus, or co-injection for mRNAs.
SELECT/SCREEN: Select or screen for mutants and verify.
VALIDATE: Genotype or sequence putative mutants to verify single or biallelic conversion.

![Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter] Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/12522/12786/sbi%2520system%2520biosciences_1633075021__00205.original__40342.1633076540.jpg?c=1)
![Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter] Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/12522/12786/sbi%20system%20biosciences_1633075021__00205.original__40342.1633076540.jpg?c=1)
![Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter] Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/12522/15578/Screenshot_2021-12-07_232449__53443.1638901540.png?c=1)
![Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter] Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/100x100/products/12522/12786/sbi%20system%20biosciences_1633075021__00205.original__40342.1633076540.jpg?c=1)
![Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter] Cas9 SmartNuclease AAVS1-gRNA Targeting Vector [EF1a promoter]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/100x100/products/12522/15578/Screenshot_2021-12-07_232449__53443.1638901540.png?c=1)