System Biosciences
CRISPY™ Master Mix for qPCR-based Gene Editing Quantitation
- SKU:
- CRISPY100A-1
- Availability:
- Usually Shipped in 5 Working Days
- Size:
- 200 rxn
- Shipping Temperature:
- Blue Ice
Description
CRISPY™ Master Mix for qPCR-based Gene Editing Quantitation. Cat# CRISPY100A. Supplier: SBI System Biosciences</

Overview
Accelerate your CRISPR/Cas9 workflows with CRISPY Master Mix, a reagent that enables fast, sensitive, and accurate quantitation of genome editing success using qPCR. Leveraging a "snapback" primer and a DNA polymerase that lacks both 5'-to-3' exoonuclease and strand displacement activity, CRISPY assays can be used to measure the percentage of successful genome edits at the target site from purified genomic DNA or cells.
The CRISPY assay offers a wealth of advantages over widely-used gel-based mismatch detection assays, including:
- Faster workflows that are complete in <1 hours with only 15 minutes of hands-on time
- Simpler workflows that involve as little as a single step, no DNA purification required
- More sensitive detection, reporting success rates as low as 1%
- More reliable quantitation based on observed Ct instead of gel imaging
- Lower sample input requirements—works on as little as 0.5 ng DNA
How It Works
The CRISPY assay relies on the use of a snapback tag that is homologous to the wildtype, unedited sequence and a DNA polymerase that lacks both 5'-to-3' exoonuclease and strand displacement activity (Figure 1).
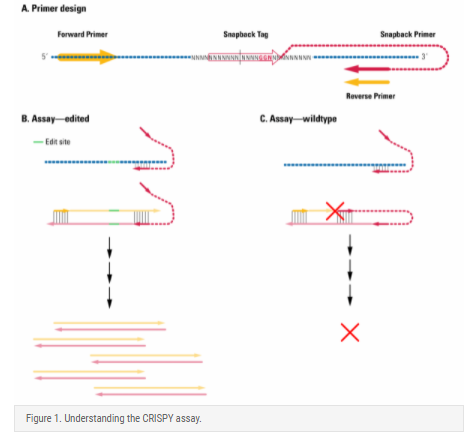
- Design forward and reverse primers (referred to here as standard primers) that will amplify across the gRNA locus. Place one of the primers within 60 nucleotides (nt) of the center of the gRNA annealing site—this will be part of your snapback primer and the other primer will be the common primer.
- Design the snapback tag by selecting 14-16 nt centered around the third or fourth base upstream of the first base in the protospacer adjacent motif (PAM) sequence. For the snapback tag, use the sequence of the wildtype, unedited sequence.
- Create the snapback primer by adding the snapback tag to the 5’-end of the designated primer (the one located within 60 nt of the center of the gRNA annealing site). If the loop formed by snapping back is shorter than 30 nt, add an optional non-homologous spacer sequence (TTTT) to ensure sufficient flexibility to form the loop.
During qPCR using common and snapback primers, only target sites that have been edited will be amplified (Figure 1B) since the snapback primer will anneal intramolecularly to primer extension products from wildtype templates, preventing amplification by the DNA polymerase which cannot displace the self-annealed snapback tag (Figure 1C).
Calculating the percentage of successful editing eventsTo calculate the percentage of successful editing events you will need to run both a control assay using the standard primers and a CRISPY assay using the common and snapback primers on both wildtype template and edited template.
Supporting Data
We edited the DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B) gene in HEK293 cells using an All-in-one Cas9 construct, isolated genomic DNA from the edited cells, and then performed a CRISPY assay to quantify editing success using standard (yellow curve) and snapback primers (red curve, Figure 2A). We also ran a CRISPY assay on DNA isolated from cells that had not been edited, again using standard (yellow curve) and Snapback primers (red curve, Figure 2B). As expected, the standard primers provided robust amplification in both edited and unedited DNA, however the snapback primers only provided appreciable amplification in the DNA isolated from edited cells. The ΔCt between the standard and snapback primers provides a direct measurement of editing success, and the difference between the shapes of the melt curves demonstrates that a deletion is present in the edited DNA (Figure 2C, boxed area).

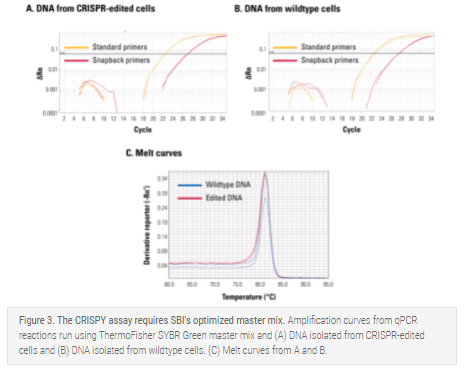
The CRISPY assay requires SBI's optimized CRISPY Master Mix
The CRISPY assay is a highly robust assay that requires the use of SBI's optimized CRISPY Master Mix—for example, ThermoFisher's SYBR Green master mix will amplify both edited (Figure 3A) and wildtype (Figure 3B) templates and provides indistinguishable melt curves (Figure 3C).
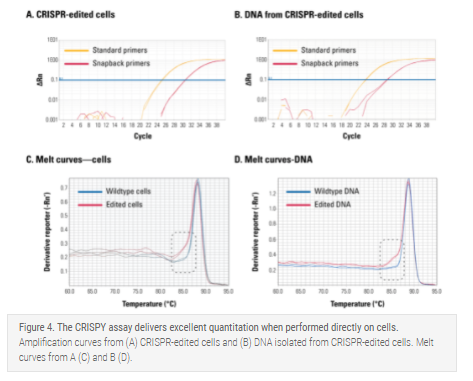
The CRISPY assay delivers excellent quantitation when performed directly on cells
To demonstrate the robustness of the CRISPY assay when using cells, we designed a new set of primers to assess the success of editing the same cells used in Figure 2. The ΔCt between standard and snapback primers in a CRISPY assay done directly on cells (Figure 4A) is virtually identical to the ΔCt in a CRISPY assay done on genomic DNA isolated from the same cells (Figure 4B), demonstrating that direct amplification of cells mirrors the results from purified DNA. This finding is also supported by comparing the melt curves performed on cells (Figure 4C) and isolated DNA (Figure 4D).