System Biosciences
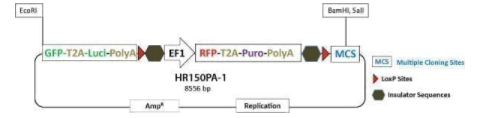
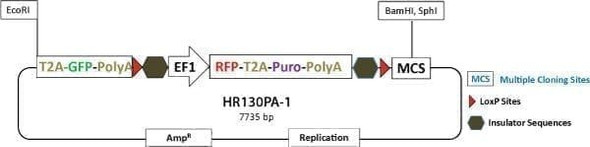
GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS]
- SKU:
- HR150PA-1
- Availability:
- Usually Shipped in 5 Working Days
- Size:
- 10 ug
- Shipping Temperature:
- Blue Ice/ Dry Ice
Description
GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS]. Cat# HR150PA. Supplier: SBI System Biosciences

Overview
Learn more about the expression of any gene using the PrecisionX™ Gene Tagging HR Targeting Vector (GFP-T2A-Luc-pA-LoxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS) which fuses GFP and co-expresses luciferase with your gene of interest. Clone your homology arms into the EcoRI site upstream of the GFP gene and into the MCS, and use puromycin selection and RFP-positive imaging to find integrants. After you’ve identified clones with your GFP-tagged gene, you can remove the selection cassette using the Cre-LoxP system (learn more about Cre-LoxP excision here).
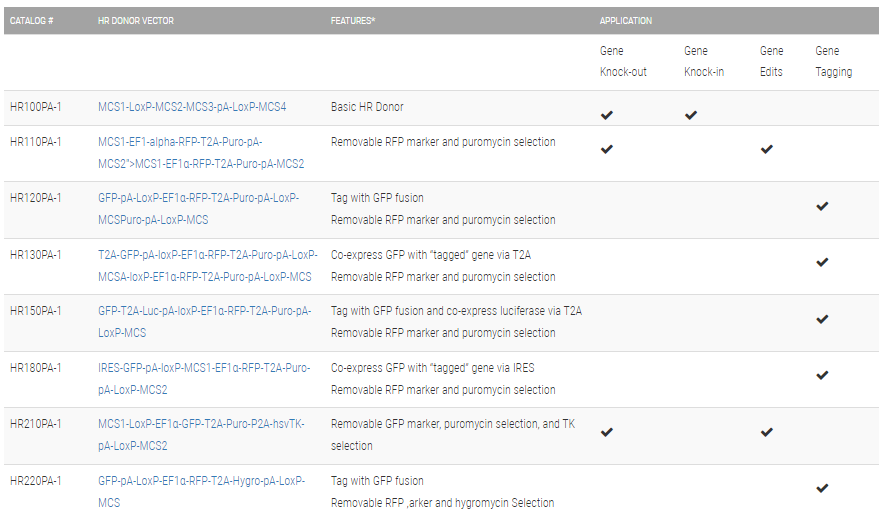
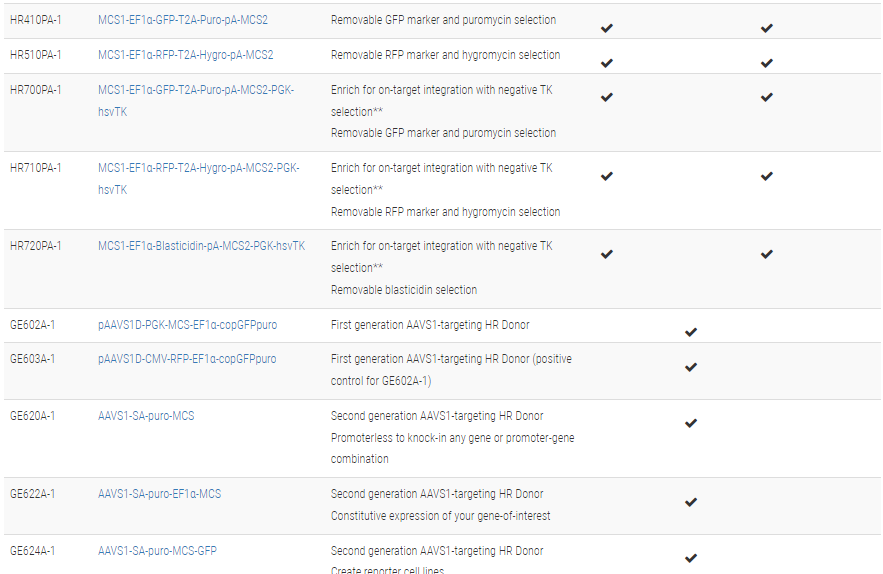
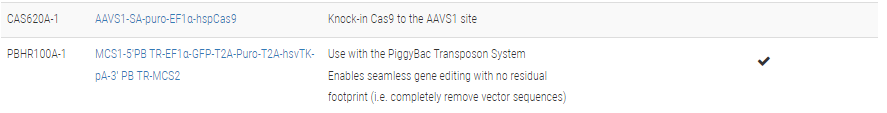
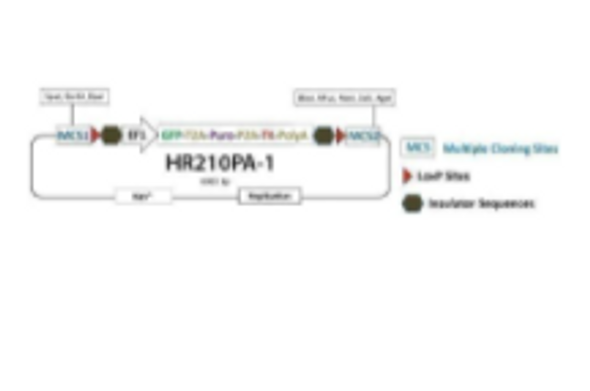
**The clever design of these HR Donors enables enrichment for on-target integration events. A PGK-hsvTK cassette is included outside of the homology arms. Because of this configuration, on-target integration that results from homologous recombination will not include the PGK-hsvTK cassette—only randomly-integrated off-target events will lead to integration of PGK-hsvTK and resulting TK activity. Therefore, TK selection will negatively select against off-target integrants. Click on any one of these vectors to see a diagram of how the negative selection works.
How It Works
At-a-glance—how to use an HR Targeting Vector to tag a gene
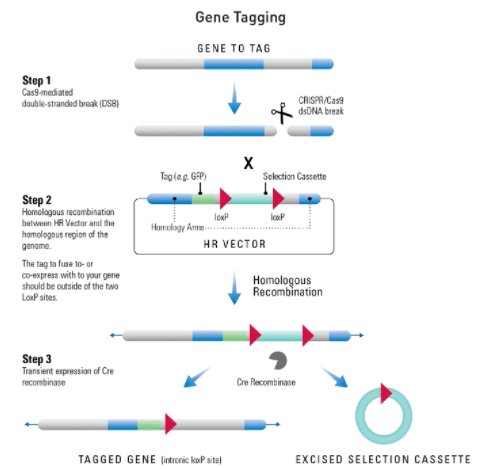
Figure 1. Tagging a gene using an HR Targeting Vector. Step 1: Cas9 creates a double-stranded break(DSB) in the genomic DNA at a site that is complimentary to the gRNA. Step 2: The DNA repair machinery is recruited to the DSB. In the presence of an HR Donor with homology to the region adjacent to the DSB (blue areas of the genomic and vector DNA) homologous recombination (HR) is favored over non-homologous end joining (NHEJ). The tag cassette you’d like to integrate into the genome (fuses GFP and co-expresses luciferase with your gene-of-interest) should lie outside of the two LoxP sites. Result: The HR event leads to insertion of the region of the HR Donor Vector between the two homology arms—your tag co-expression cassette is integrated to the C-terminus of your gene-of-interest and your selection cassette is integrated into the gene. To remove the selection cassette (leaving behind the tag and a single LoxP site), transiently express Cre recombinase.
Genome engineering with CRISPR/Cas9
For general guidance on using CRISPR/Cas9 technology for genome engineering, including the design of HR Targeting Vectors, take a look at our CRISPR/Cas9 tutorials as well as the following application notes:
CRISPR/Cas9 Gene Knock-Out Application Note (PDF) »
CRISPR/Cas9 Gene Editing Application Note (PDF) »
CRISPR/Cas9 Gene Tagging Application Note (PDF) »

![GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS] GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/12680/12945/sbi%2520system%2520biosciences_1633075021__00205.original__65000.1633123753.jpg?c=1)
![GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS] GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/12680/12945/sbi%20system%20biosciences_1633075021__00205.original__65000.1633123753.jpg?c=1)
![GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS] GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/12680/15650/Screenshot_2021-12-09_224538__37530.1639071973.png?c=1)
![GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS] GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/100x100/products/12680/12945/sbi%20system%20biosciences_1633075021__00205.original__65000.1633123753.jpg?c=1)
![GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS] GFP-T2A-Luciferase Co-Expression HR Targeting Vector [GFP-T2A-Luc-pA-loxP-EF1α-RFP-T2A-Puro-pA-LoxP-MCS]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/100x100/products/12680/15650/Screenshot_2021-12-09_224538__37530.1639071973.png?c=1)





![GFP-Fusion HR Targeting Vector [GFP-pA-LoxP-EF1a-RFP-T2A-Hygro-pA-LoxP-MCS] GFP-Fusion HR Targeting Vector [GFP-pA-LoxP-EF1a-RFP-T2A-Hygro-pA-LoxP-MCS]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/590x590/products/12678/12943/sbi%2520system%2520biosciences_1633075021__00205.original__31669.1633076565.jpg?c=1)
![GFP-Fusion HR Targeting Vector [GFP-pA-LoxP-EF1a-RFP-T2A-Hygro-pA-LoxP-MCS] GFP-Fusion HR Targeting Vector [GFP-pA-LoxP-EF1a-RFP-T2A-Hygro-pA-LoxP-MCS]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/590x590/products/12678/15648/Screenshot_2021-12-09_223452__34777.1639071351.png?c=1)