System Biosciences
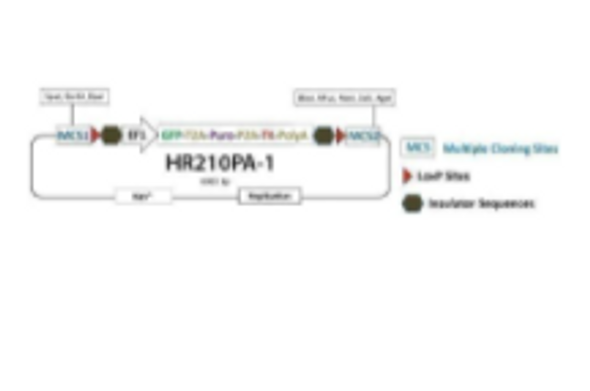
piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correction
- SKU:
- PBHR100A-1
- Availability:
- Usually Shipped in 5 Working Days
- Size:
- 10 ug
- Shipping Temperature:
- Blue Ice/ Dry Ice
- Qualifications:
- Requires signed license from commercial customers
Description
piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correction. Cat# PBHR100A. Supplier: SBI System Biosciences
Products

Overview
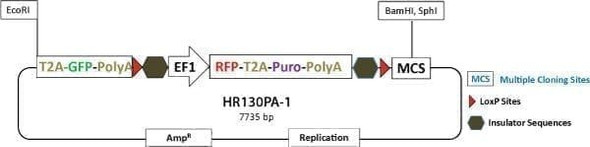
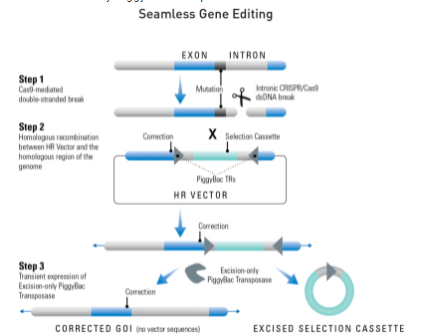
Use the PiggyBac Gene Editing HR Targeting Vector (MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2) to get seamless gene editing—leave no trace of vector sequences behind.
This special HR Donor works with the Excision-only PiggyBac Transposase instead of the Cre-LoxP system. Simply proceed with your CRISPR/Cas9 gene editing as usual—clone your homology arms into MCS1 and MCS2, use dual GFP and puromycin selection to find integrants, and enrich for on-target events using negative thymidine kinase (TK) selection (Figure 1).

Why use an HR targeting vector?
Even though gene knock-outs can result from DSBs caused by Cas9 alone, SBI recommends the use of HR targeting vectors (also called HR donor vectors) for more efficient and precise mutation. HR donors can supply elements for positive or negative selection ensuring easier identification of successful mutation events. In addition, HR donors can include up to 6-8 kb of open reading frame for gene knock-ins or tagging, and, when small mutations are included in either 5’ or 3’ homology arms, can make specific, targeted gene edits.
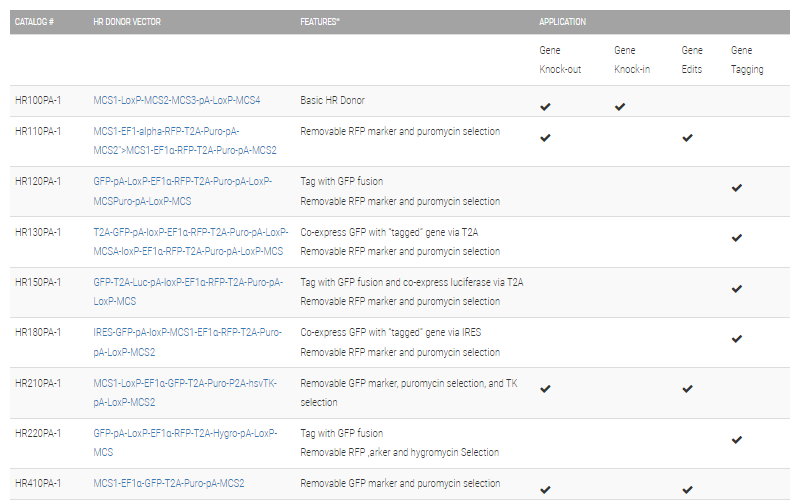
Choose the right HR Targeting Vector for your project
How It Works
1 Review
-
superb
PIGGYBAC-HR WITH GFP+PURO MARKERS AND TK SELECTION was a superb product and is recommended.

![piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/14421/14689/sbi%2520system%2520biosciences_1633075021__00205.original__16600.1633076840.jpg?c=1)
![piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/14421/14689/sbi%20system%20biosciences_1633075021__00205.original__16600.1633076840.jpg?c=1)
![piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/608x608/products/14421/29947/PBHR100A-1__86845.1640081714.png?c=1)
![piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/100x100/products/14421/14689/sbi%20system%20biosciences_1633075021__00205.original__16600.1633076840.jpg?c=1)
![piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio piggyBac-HR with GFP+Puro markers and TK selection [MCS1-5'PB TR-EF1α-GFP-T2A-Puro-T2A-hsvTK-pA-3' PB TR-MCS2] for Gene Correctio](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/100x100/products/14421/29947/PBHR100A-1__86845.1640081714.png?c=1)

![Gene Knock-Out HR Targeting Vector [MCS1-EF1a-GFP-T2A-Puro-pA-MCS2] Gene Knock-Out HR Targeting Vector [MCS1-EF1a-GFP-T2A-Puro-pA-MCS2]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/590x590/products/12671/12936/sbi%2520system%2520biosciences_1633075021__00205.original__02911.1633076564.jpg?c=1)
![Gene Knock-Out HR Targeting Vector [MCS1-EF1a-GFP-T2A-Puro-pA-MCS2] Gene Knock-Out HR Targeting Vector [MCS1-EF1a-GFP-T2A-Puro-pA-MCS2]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/590x590/products/12671/15641/Screenshot_2021-12-09_212928__74392.1639067413.png?c=1)
![Gene Knock-Out HR Targeting Vector [MCS1-EF1α-RFP-T2A-Puro-pA-MCS2] Gene Knock-Out HR Targeting Vector [MCS1-EF1α-RFP-T2A-Puro-pA-MCS2]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/590x590/products/12673/12938/sbi%2520system%2520biosciences_1633075021__00205.original__03121.1633123705.jpg?c=1)
![Gene Knock-Out HR Targeting Vector [MCS1-EF1α-RFP-T2A-Puro-pA-MCS2] Gene Knock-Out HR Targeting Vector [MCS1-EF1α-RFP-T2A-Puro-pA-MCS2]](https://cdn11.bigcommerce.com/s-i3n9sxgjum/images/stencil/590x590/products/12673/15643/Screenshot_2021-12-09_214631__77660.1640262373.png?c=1)